Drug discovery is notorious for its low success rates [1]. Despite best research efforts, the majority of drugs fail at early stages of development, even before they enter clinical trials. This phenomenon stems from the inherent complexity of biological systems and our poor understanding of human diseases. To improve that understanding, swaths of data have been generated in recent years. Still, data does not easily translate into knowledge or actionable insights. Here we explore how approaches from the recommendation system domain could help scientists comprehend the ever-growing amount of biomedical facts. The aim of these efforts is to make better drug development decisions, which ultimately result in safe and efficient treatments for patients.
Recommendation systems are well established in e-commerce, streaming and social media platforms, however in the biomedical domain their usage is limited to a few recent studies [2, 3, 4, 5]. Direct transfer of classic recommendation approaches to the biomedical domain is not trivial. Specifics of the problem space impose numerous challenges for a recommendation system practitioner, to name a few:
-
an elementary unit of recommendation is not a simple self-contained item (like a song or a product), but rather a research direction accompanied by a biologically sound hypothesis;
-
ultimate validation of recommendations is complex and often requires expensive and time-consuming laboratory experiments;
-
unlike traditional applications, in a biomedical setting both implicit and explicit feedback is at best scarce, making it harder to tune and train models;
-
ground truths are scarce which renders training challenging;
-
context matters, in a sense that each diseases is a heterogeneous and complex molecular phenomenon; depending on what aspect of a disease we want to focus on, different recommendations are expected;
-
due to the high cost associated with accepting a recommendation, an increased emphasis is placed on explainability and exposing causal reasoning paths behind a recommendation;
-
personalized recommendations approaches are not applicable: in biology or drug discovery decisions are rarely taken by a single user, but rather a team of experts each with different bias;
-
previous literature significantly biases users decisions. Users tend to favour known entities, rather than opting for completely novel recommendations (e.g genes residing in the ‘dark matter’ of the human genome). Adding to this inherent bias, the design of experimental validations is more challenging where less prior knowledge is available.
Regardless of the challenges, the adoption of recommendation systems presents numerous opportunities to support and accelerate drug discovery. Even a slight increase in success rate of drug pipelines will result in a vast number of patients gaining access to safe and effective treatments. Recommendation systems could play a leading role in this process.
Adding context to experimental data is one class of problems that could benefit from recommenders. In this process new data is integrated with prior evidence to produce a new hypothesis. In a typical scenario, thousands of genes need to be ranked by their relevance to a disease given new and existing data. As a case study we focused on finding out why some lung cancer patients develop resistance to treatments. Current protocol to find resistance markers starts with high-throughput genomic screens resulting in an initial list of potential gene candidates, followed by tedious manual curation by several experts to reduce the list to a manageable number for further follow-up.
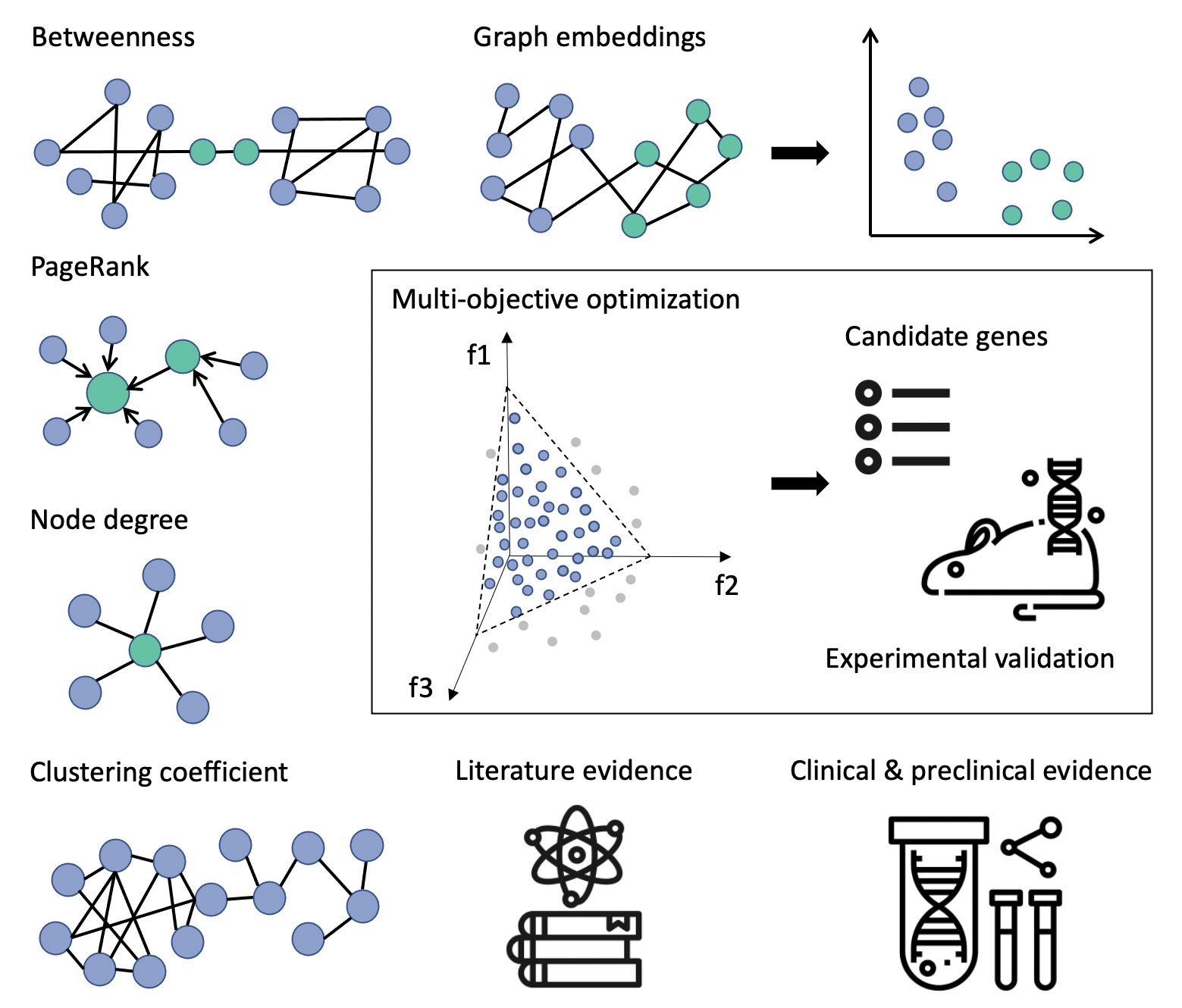
Figure 1. Recommendation system takes into account diverse types of evidence to suggest promising genes driving drug resistance in lung cancer patients. Recommended genes are experimentally validated in disease models.
To find resistance markers faster and to reduce bias we built a hybrid recommendation system on top of a heterogeneous biomedical knowledge graph [6]. In the absence of continuous feedback and training data, we approached recommendations as a multi-objective optimization problem [7]. Genes were ranked by trading off diverse types of evidence that link them to potential mechanisms of resistance in lung cancer. We used a knowledge graph as the primary source of features, so that the relevance of a gene could be expressed via properties of a graph. Our hybrid feature set also included clinical and pre-clinical data as well as metrics of literature support obtained with natural language processing techniques. This hybrid approach helped to identify novel resistance mechanisms that could have been overlooked by experts due to inherent bias or limited integration of data. Most importantly, our method reduced the time required to prioritise resistance markers from months to minutes and became a standard procedure for processing genomic screens.
Another class of problems exists around target identification tasks. The idea here is to find a molecular target, often a gene or a protein, that could be modulated with a drug to treat a disease. As the number of potential targets is large, the search space can be reduced using network propagation on a dedicated subgraph that captures the functional relationship between genes. This approach also requires a set of seed genes, defined based on high confidence associations with diseases. Disease preferences are then propagated through the network resulting in a preference distribution for the complete set of genes which is used to reduce the search space.
In contrast to adding context to experiments, a considerable amount of training data is available to support target identification. For instance, both successful and failed clinical trials can act as a useful source of data for target identification. Such a setting warrants use of supervised recommendation systems. A supervised approach, however introduces another machine learning hurdle — trust. Since supervised models are typically "black boxes", their quality must be ascertained indirectly, for example using train-test split and estimating model’s performance on the test set. Such quantitative performance metrics often are of little value to a biological expert looking for relevant gene targets. Instead, experts instinctively assess model quality by checking if a list of recommendations contains a handful of expected genes [8].
To simultaneously use biologists’ intuitions as training data, while avoiding an overly optimistic trust in model output, we used an ensemble modeling approach. We partitioned training data among multiple models such that each available training gene was omitted from one model’s training data. The model was then permitted to assess this previously unseen gene in constructing its final list of recommendations, while training genes were removed from consideration. Each model therefore produced a list of recommendations based on an incomplete set of genes. A final set of recommendations was then constructed by collating each individual model’s output list. Because these output lists were constructed with biologist input through supervised training, biologists placed a higher degree of trust in the recommendations. This allowed roughly two dozen genes to be fast-tracked for manual assessment and experimental screening.
In summary, accumulation of large amounts of biomedical data coupled with a need to comprehend and reason about it makes drug discovery an attractive field to apply recommendation techniques. Specifics of the problem space and complexity of biological systems call for efficient recommendation solutions that could operate in unsupervised or weakly supervised settings. At the same time, a strong emphasis on explainability is essential to gain trust of biomedical experts.
We thank Gavin Edwards, Andrej Lamov, Krisha Bulusu, Dimitris Polychronopoulos and Benedek Rozemberczki for technical and scientific support.
References
[1] Paul Morgan et al (2018). Impact of a five-dimensional framework on R\&D productivity at AstraZeneca. Nature reviews Drug discovery, 17(3), 167 - 181.
[2] Ozsoy et al (2018). Realizing drug repositioning by adapting a recommendation system to handle the process. BMC Bioinformatics, 19(1), 136.
[3] Frainay et al (2019). MetaboRank: network-based recommendation system to interpret and enrich metabolomics results. Bioinformatics, 35(2), 274 - 283.
[4] Suphavilai et al (2018). Predicting cancer drug response using a recommender system. Bioinformatics, 34(22), 3907 - 3914.
[5] Radivojevic et al (2020). A machine learning Automated Recommendation Tool for synthetic biology. Nature Communications, 11(1), 1 - 14.
[6] Geleta & Nikilov (in preparation). Biological Insights Knowledge Graph.
[7] Gogleva et al (2021). Knowledge Graph-based Recommendation Framework Identifies Novel Drivers of Resistance in EGFR mutant Non-small Cell Lung Cancer. bioRxiv 2021.07.23.453506; doi: https://doi.org/10.1101/2021.07.23.453506
[8] Zhang et al (2018). How Well do Offline and Online Evaluation Metrics Measure User Satisfaction in Web Image Search? The 41st International ACM SIGIR Conference on Research \& Development in Information Retrieval, 615 - 624.
